Connections in the Human Structural Connectome
Introduction
From the human structural connectome, the authors'\(^{[1]}\) attempt to extract architectural features using diffusion spectrum imaging DSI and encode the data required into triplcate as undirected, weighted network. They do so to capture as much information about possible paths which transmit as human process and preform complex behaviors.
Persistent Homology in Neural Networks
Our interest is in isolating mesoscale structural features, specifically complex minimal cycles in weighted neural networks. First, we will identify cliques or brain regions that rapidly and effectively share information. Formally, let a \((k+1)\)-clique of graph \(G\) be a set of \((k+1)\) pairwise edges in \(G\). A subgraph of a clique must be a clique itself of a lower degree, which is called a face.
To continue.
Local Neighborhoods in Strucutral Brain Networks
Clique Features of Local Neighborhoods
Clique Feature Representation
Neurological data can be summairzed as a network of corresponding node brain regions with weighted edges that is represented as the density of white matter steamlines reconstructed between corresepodning nodes.
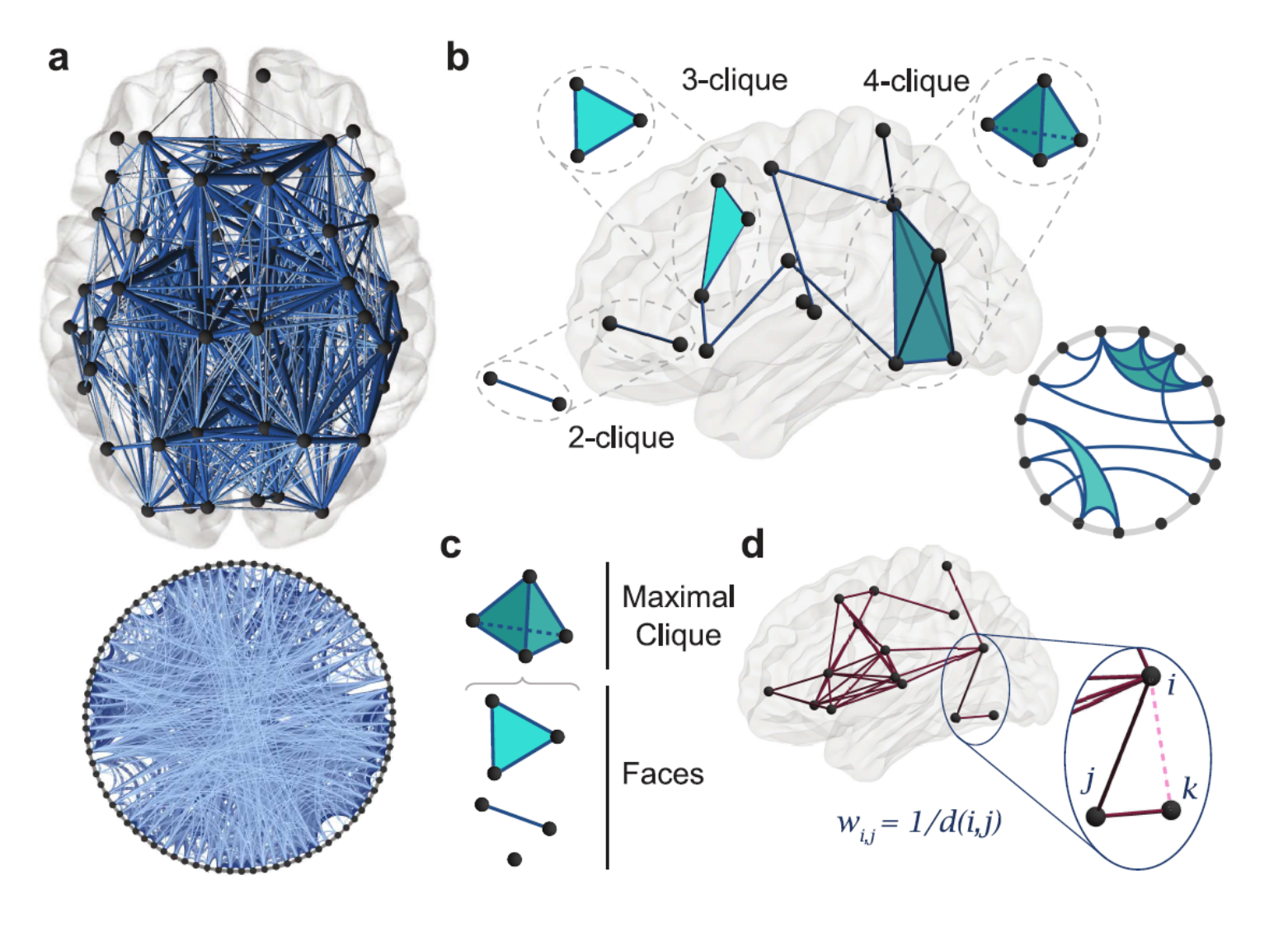
Image Source: See Reference [1]
In the above figure, the authors present a group-averaged network that has an edge density \(\rho=0.25\). For values examined as null-models with minimally Euclidean invasive wired networks, weights assigning edge weights inverse between brain regions can be assigned.
Appendix
Diffusion Specturm Imaging DSI
Diffusion Spectrum Imaging (DSI) is an advanced magnetic resonance imaging (MRI) technique used in neuroimaging to study complex tissue microstructures, particularly in the brain.